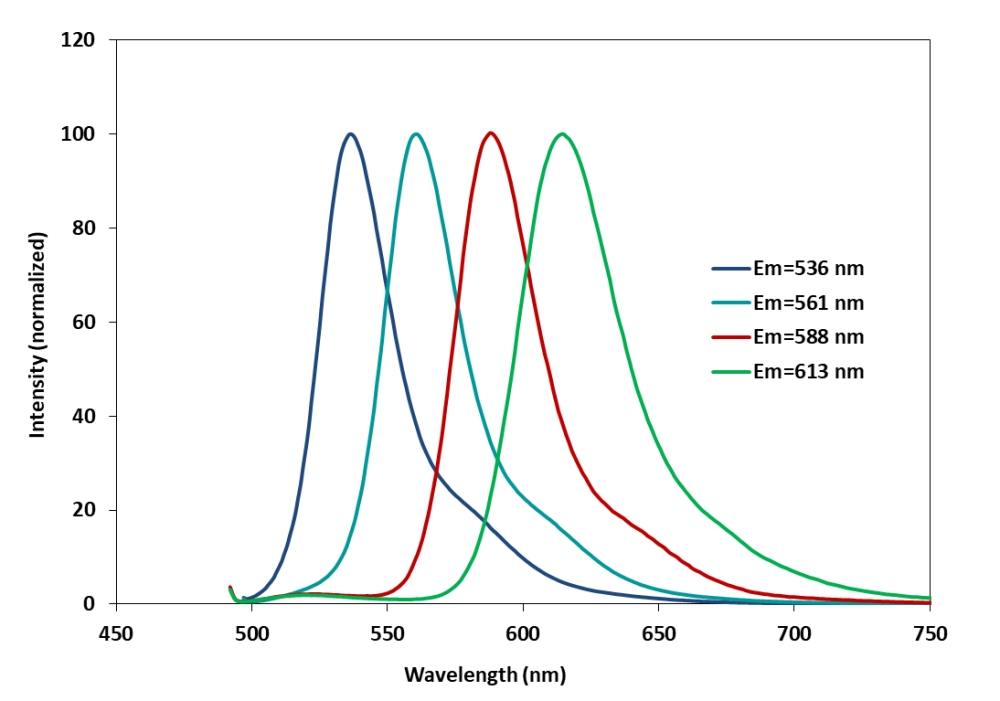
MagaDye 588-ddTTP
 |
货号 |
17061 |
存储条件 |
在零下15度以下保存, 避免光照 |
| 规格 |
5 nmoles |
价格 |
11988 |
| Ex (nm) |
498 |
Em (nm) |
588 |
| 分子量 |
~1900 |
溶剂 |
Water |
| 产品详细介绍 |
简要概述
产品基本信息
货号:17061
产品名称:MagaDye 588-ddTTP
规格:5nmoles
储存条件:保存在冰箱-15℃干燥
保质期:12个月
产品物理化学光谱特性
Ex(nm):498
Em(nm):588
吸收(nm):498
产品介绍
Sanger测序,也称为链终止法,是一种基于DNA聚合酶选择性掺入链终止双脱氧核苷酸(ddNTPs)的DNA测序技术。虽然新的NGS技术由于其较高的通量能力和较低的每份样品成本而在临床研究实验室中变得很普遍,但Sanger测序仍具有99.99%的准确度。四种不同的荧光ddNTP(标记为BigDye®,BigDye®是ThermoFisher的商标)是执行Sanger测序的关键成分。MagaDye 588-ddTTP等同于BigDye dROX,具有几乎相同的光谱。金畔生物是AAT Bioquest的中国代理商,为您提供最优质的MagaDye 588-ddTTP。
点击查看光谱
参考文献
A novel gross deletion and breakpoint junction sequence analysis of ATP7B in a Chinese family with Wilson disease using next‑generation sequencing and Sanger sequencing.
Authors: Liu, Wei-Liang and Li, Fang and Liu, Lu and Chen, Wei and He, Zhi-Xu and Gu, Hao and Ai, Rong
Journal: Molecular medicine reports (2020): 517-523
Concurrent Cultivation of Mycobacterium avium and Mycobacterium intracellulare Identified by a Single Sanger Sequencing of the 16S Gene.
Authors: Han, Xiang Y and Golshan, Mohammad A and Bowman, Christopher J
Journal: Journal of clinical microbiology (2020)
Detection of TERT promoter mutation in serum cell-free DNA using wild-type blocking PCR combined with Sanger sequencing in hepatocellular carcinoma.
Authors: Akuta, Norio and Suzuki, Fumitaka and Kobayashi, Mariko and Fujiyama, Shunichiro and Kawamura, Yusuke and Sezaki, Hitomi and Hosaka, Tetsuya and Kobayashi, Masahiro and Saitoh, Satoshi and Arase, Yasuji and Ikeda, Kenji and Suzuki, Yoshiyuki and Kumada, Hiromitsu
Journal: Journal of medical virology (2020)
Guidelines for Sanger sequencing and molecular assay monitoring.
Authors: Crossley, Beate M and Bai, Jianfa and Glaser, Amy and Maes, Roger and Porter, Elizabeth and Killian, Mary Lea and Clement, Travis and Toohey-Kurth, Kathy
Journal: Journal of veterinary diagnostic investigation : official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc (2020): 1040638720905833
Rapid, Inexpensive Measurement of Synthetic Bacterial Community Composition by Sanger Sequencing of Amplicon Mixtures.
Authors: Cermak, Nathan and Datta, Manoshi Sen and Conwill, Arolyn
Journal: iScience (2020): 100915
Shall I trust the report? Variable performance of Sanger sequencing revealed by deep sequencing on HIV drug resistance mutation detection.
Authors: Chen, Nan-Yu and Kao, Shu-Wei and Liu, Zhuo-Hao and Wu, Ting-Shu and Tsai, Chia-Lung and Lin, Hsi-Hsun and Wong, Wing-Wai and Chang, Yea-Yuan and Chen, Shu-Sheng and Ku, Stephane Wen-Wei
Journal: International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases (2020): 182-191
Update on molecular companion diagnostics – a future in personalized medicine beyond Sanger sequencing.
Authors: Campbell, Michelle Renee
Journal: Expert review of molecular diagnostics (2020)
BEAT: A Python Program to Quantify Base Editing from Sanger Sequencing.
Authors: Xu, Li and Liu, Yakun and Han, Renzhi
Journal: The CRISPR journal (2019): 223-229
Characterization and Clinical Significance of Natural Variability in Hepatitis B Virus Reverse Transcriptase in Treatment-Naive Chinese Patients by Sanger Sequencing and Next-Generation Sequencing.
Authors: Fu, Ya and Zeng, Yongbin and Chen, Tianbin and Chen, Huijuan and Lin, Ni and Lin, Jinpiao and Liu, Xiaofeng and Huang, Er and Wu, Songhang and Wu, Shu and Xu, Siyi and Wang, Long and Ou, Qishui
Journal: Journal of clinical microbiology (2019)
Comparison of Sanger sequencing for hepatitis C virus genotyping with a commercial line probe assay in a tertiary hospital.
Authors: Goletti, Sylvie and Zuyten, Siméon and Goeminne, Léonie and Verhofstede, Chris and Rodriguez-Villalobos, Hector and Bodeus, Monique and Stärkel, Peter and Horsmans, Yves and Kabamba-Mukadi, Benoît
Journal: BMC infectious diseases (2019): 738
说明书
MagaDye 588-ddTTP.pdf